Modeling metabolism
Metabolism describes the conversion of nutrients into cellular building blocks and energy. Mathematical models - ranging form single metabolic pathways such as glycolysis to genome-scale metabolic networks - allow to unravel underlying regulatory mechanisms as well as to understand systemic features not accessible to experimental analyses. In addition, those models can predict essential features of the investigated systems, thus generating hypotheses for experimental validation and future research. |
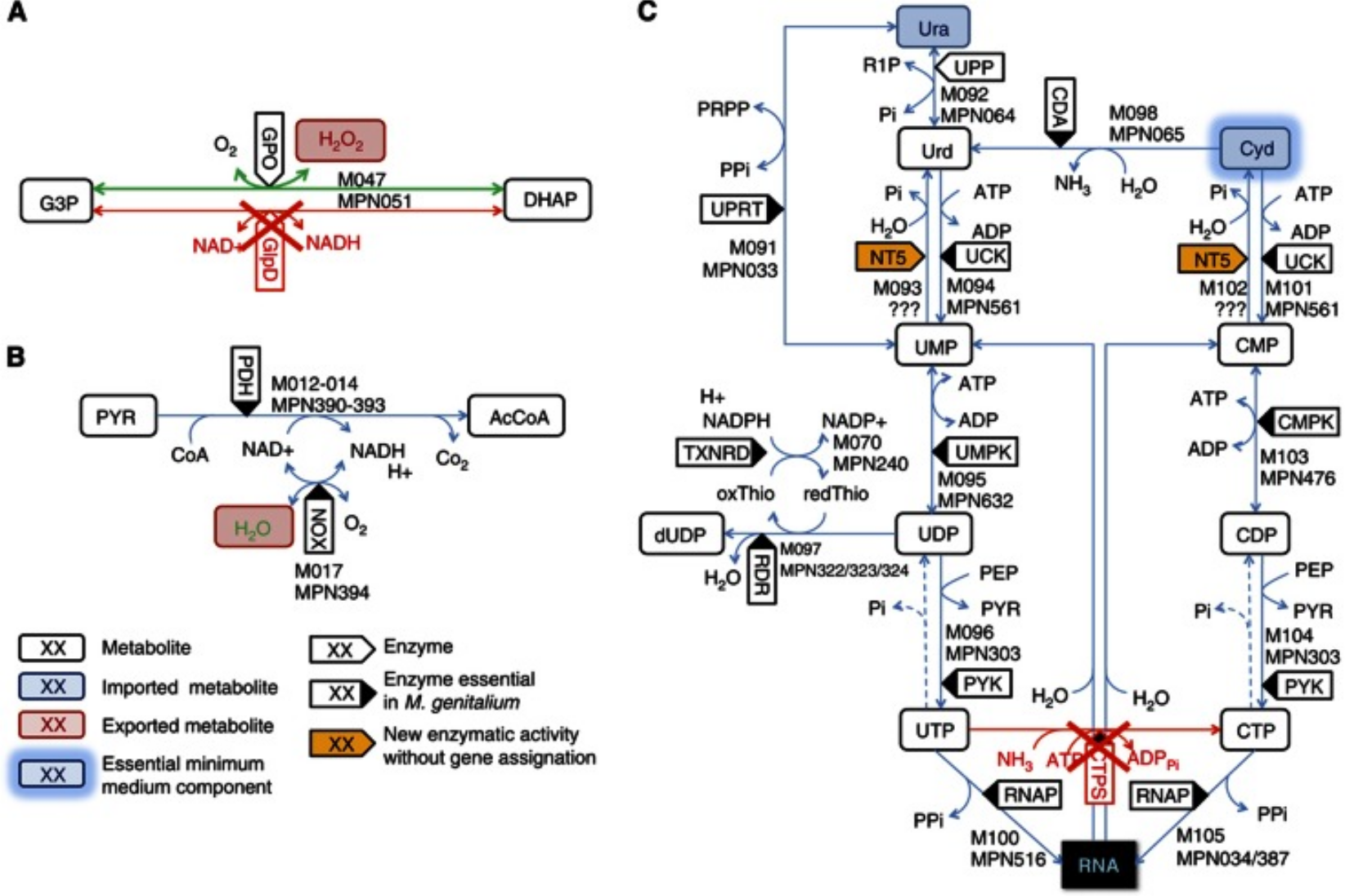
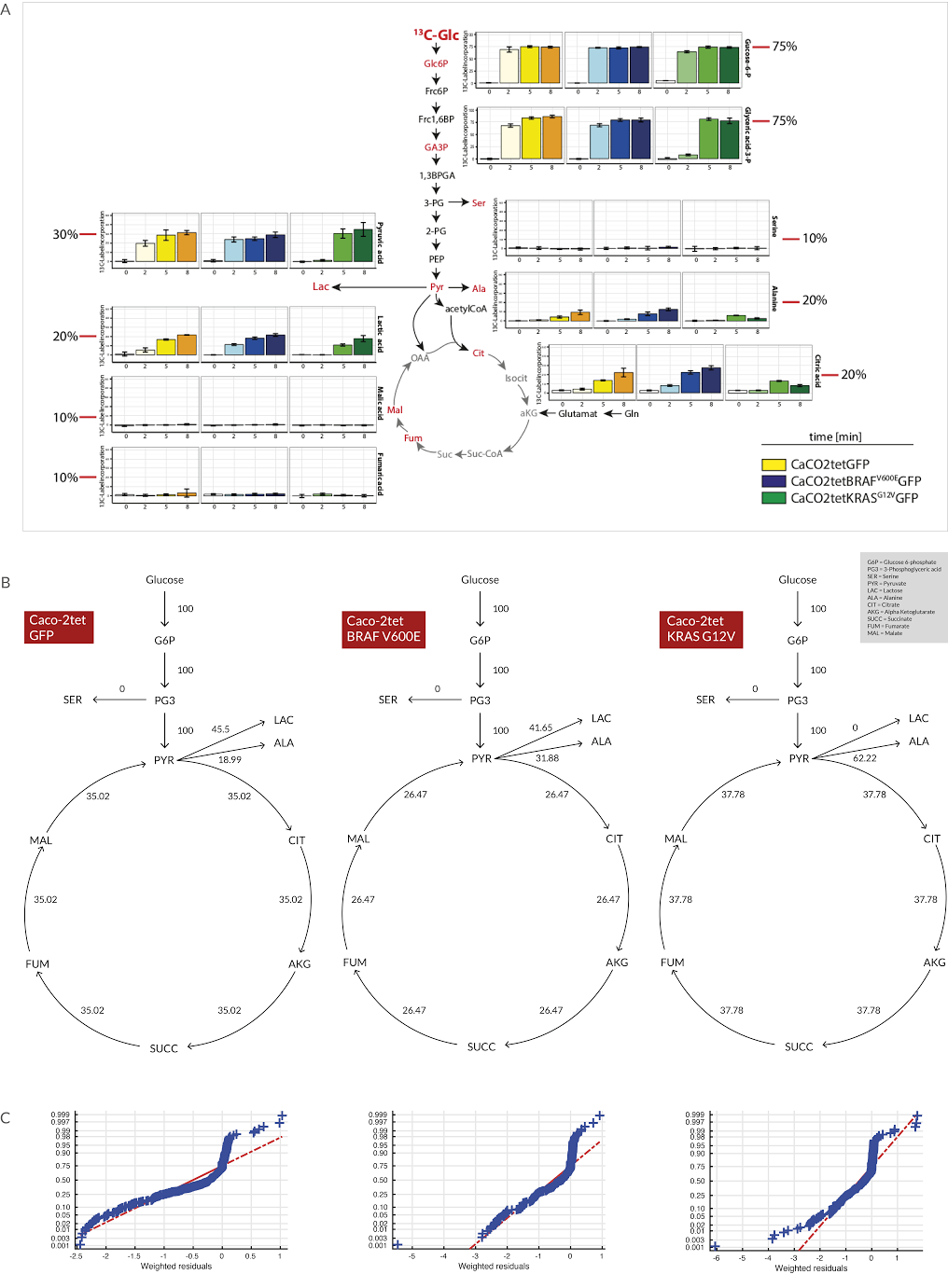
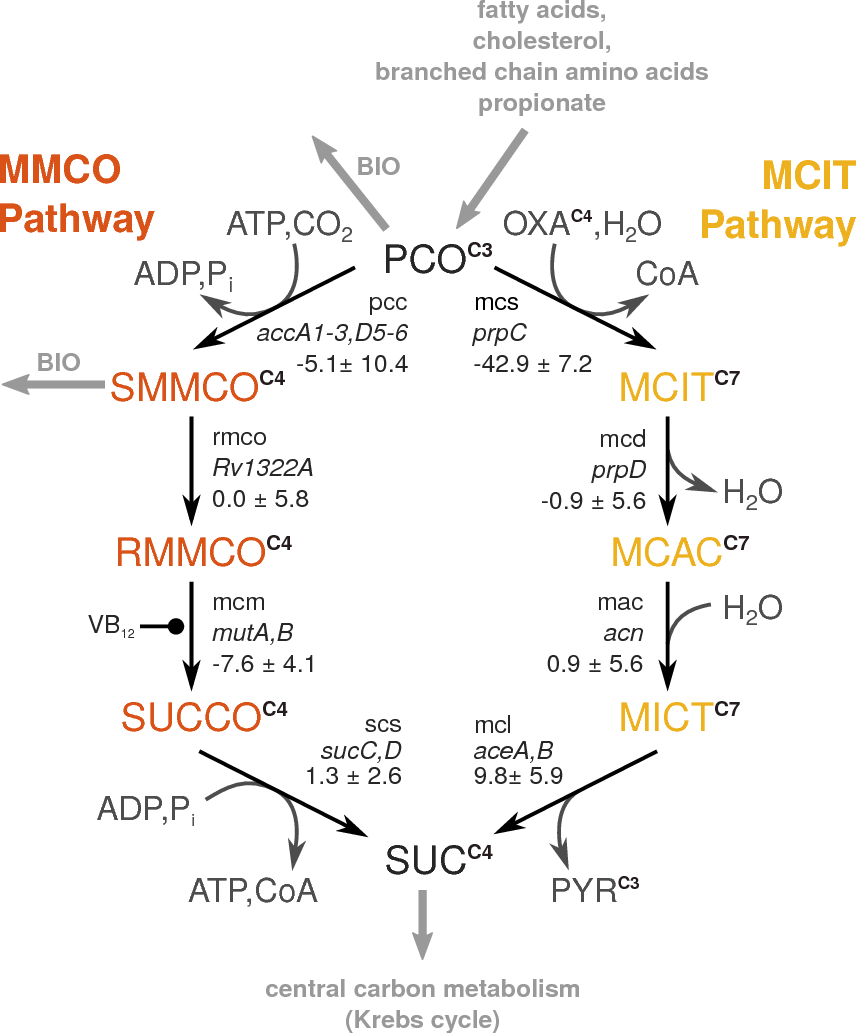
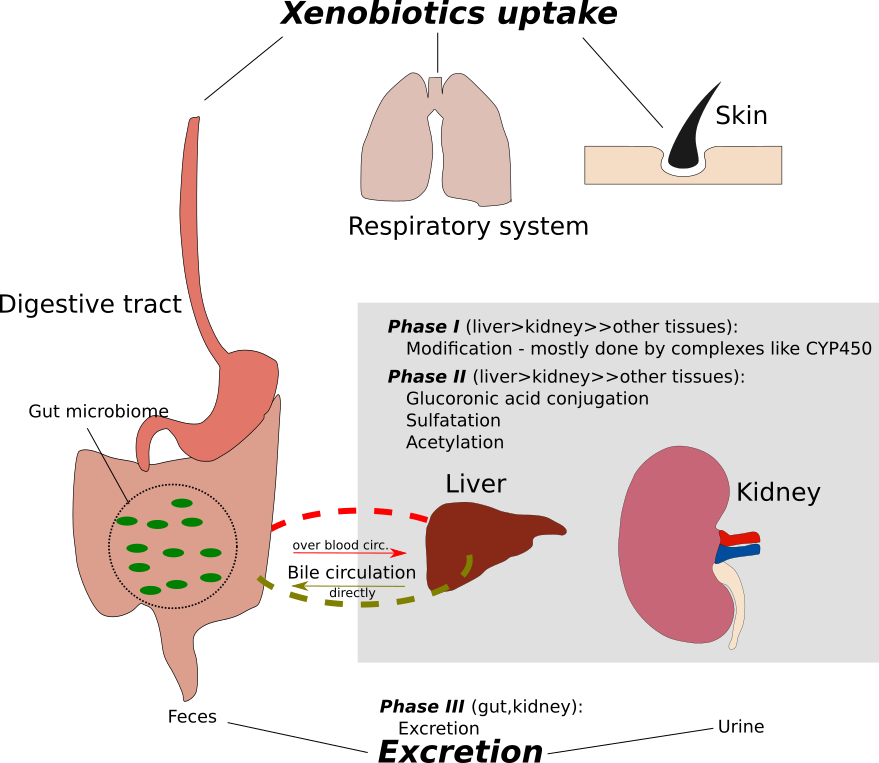
Image from Wodke et al., Mol Sys Biol (2013), CC BY 4.0 license |
PEOPLE
PROJECTS
SELECTED PUBLICATIONS
T Lubitz, M Schulz, E Klipp and W Liebermeister.
Parameter balancing in kinetic models of cell metabolism.
J Phys. Chem. B 114 (49):16298–16303, December 2010.
URLN J Stanford, T Lubitz, K Smallbone, E Klipp, P Mendes and W Liebermeister.
Systematic construction of kinetic models from genome-scale metabolic networks.
PLoS ONE 8 (11):e79195, 2013.
URLJ A H Wodke, J Puchałka, M Lluch-Senar, J Marcos, E Yus, M Godinho, R Gutiérrez-Gallego, V A P Martins dos Santos, L Serrano, E Klipp and T Maier.
Dissecting the energy metabolism in Mycoplasma pneumoniae through genome-scale metabolic modeling.
Mol. Syst. Biol. 9:653, 2013.
URLT Lubitz, J Hahn, F T Bergmann, E Noor, E Klipp and W Liebermeister.
SBtab: a flexible table format for data exchange in systems biology.
Bioinformatics 32 (16):2559–2561, 2016.
URLJ Pauling and E Klipp.
Computational lipidomics and lipid bioinformatics: filling In the blanks.
J. Integr. Bioinform. 13 (1):299, 2016.
URL
FUNDINGBMBF Consejo Nacional de Ciencia y Tecnología (CONACyT) DAAD EU - Marie Curie Research Fellowship Programme EU - Sixth Framework Programme EU - Seventh Framework Programme DFG |
COLLABORATIONSFritsche, Raphaela & Kempa, Stefan, MDC Berlin (Germany) Hampe, Jochen, Universitätsklinikum Carl Gustav Carus, Dresden (Germany) Liebermeister, Wolfram , Charite Berlin (Germany) Sauer group, ETH Zurich (Switzerland) Shevchenko, Andrej, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden (Germany) LiSyM network, consortium of research groups (Germany)SysToxChip consortium Department of biochemistry at Charité, Berlin BioSys (department: MaIAGE - Mathématiques et Informatique Appliquées du Génome à l'Environnement) at INRA (Institut national de la recherche agronomique) in Jouy-en-Josas, France |