As a part of the Klipp Lab’s yeast cell modelling efforts, our lab deploys an array of imaging techniques, such as brightfield imaging and fluorescence imaging, in order to parameterize cellular processes like cell-cycle propagation.
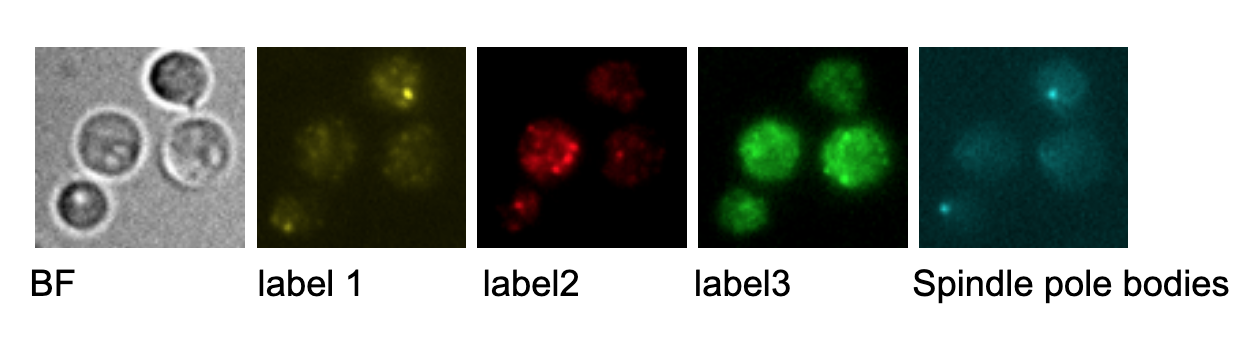
In this project, Multi-Sequential Single-Molecule Fluorescence in Situ Hybridization (MuSeqFISH) is used to visualize single mRNA molecules of genes relevant to the cell cycle regulation and determine their temporal correlation. Visualization of single mRNA molecules is facilitated through attached fluorescent labeled DNA probes that are specific for the target genes of interest. After imaging the cells on a widefield microscope the labels are washed away and the cells are labeled again with probes for different targets. Repetition of this procedure allows to gain mRNA numbers for up to 21 genes of the same cell. The obtained microscopic images are subsequently processed and analyzed.
Your responsibility will be, depending on both your personal interests and the task at hand, to perform experiments using an established protocol, including cultivation and modification of yeast cultures, microscopic imaging, image analysis, and statistical analysis. Your work will contribute to understanding the choreography of relevant transcripts during the cell cycle and will be embedded in a larger project that aims to understand whole cell dynamics of S. cerevisiae.
Topics and Keywords
S.cerevisiae, mRNA, cell-cycle, Fluorescence Microscopy
Tasks and Milestones
1. Yeast culturing and yeast modification
2. Quantitaive microscopic imaging
3. Statistical analysis
5. Report writing
Literature
Amoussouvi A, Teufel L, Reis M, et al. Transcriptional timing and noise of yeast cell cycle regulators-a single cell and single molecule approach. NPJ Syst Biol Appl. 2018;4:17. Published 2018 May 21. doi:10.1038/s41540-018-0053-4
Haralampiev, I., Prisner, S., Nitzan, M. et al. Selective flexible packaging pathways of the segmented genome of influenza A virus. Nat Commun 11, 4355 (2020). https://doi.org/10.1038/s41467-020-18108-1

