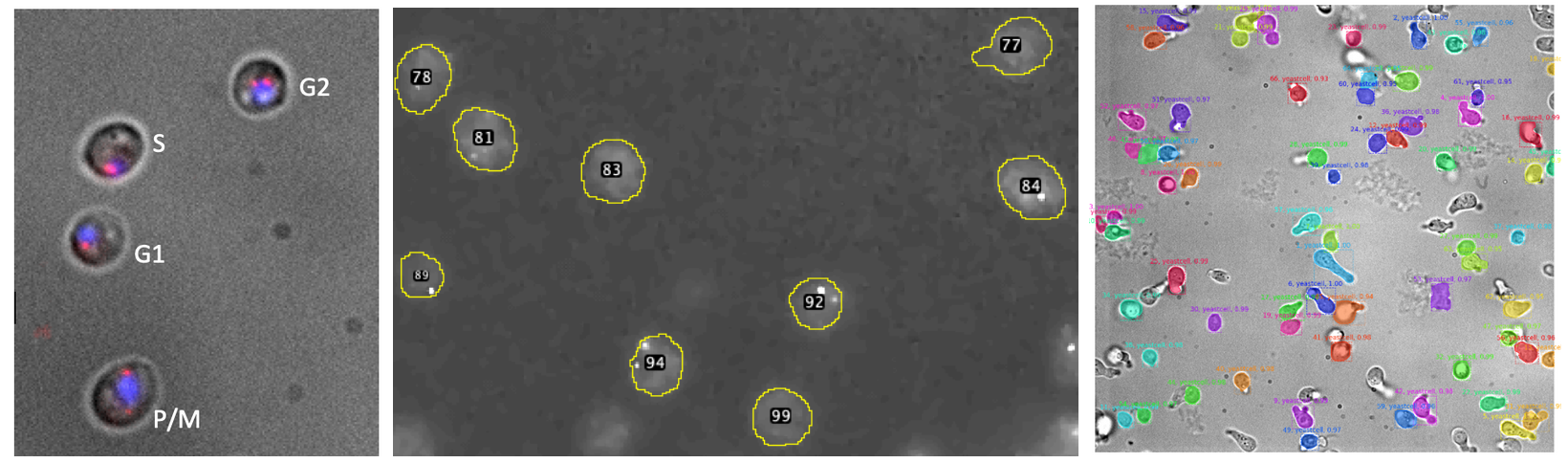
As a part of the Klipp Lab’s yeast cell modelling efforts, our lab deploys an array of imaging techniques, such as brightfield imaging and fluorescence imaging, in order to parameterize cellular processes like cell-cycle propagation.
One of the lab’s main current projects is exploring and parameterizing the expression of cell-cycle phase-specific proteins called cyclins in unsynchronized yeast cells. In order to resolve the cyclin expression levels in each cell cycle phase in single cells, single cell cycle relevant mRNAs are fluorescently labeled and imaged using fluorescence and wide field microscopy. This project revolves around further developing a pre-existing deep learning cell segmentation and morphological classifier so as to learn from both the morphology of single cells as well as the presence and colocalization of relevant fluorescence markers in order to classify cellular imaging data, thereby enabling automated cell-cycle phase assignment.
Your task will therefore be to develop an all-in-one machine learning based solution for automatic assignment cell-cycle phases from fluorescence and brigh-field images. Your work will contribute to understanding the choreography of relevant transcripts during the cell cycle and will be embedded in a larger project that aims to understand whole cell dynamics of S. cerevisiae.
Topics and Keywords
S.cerevisiae, mRNA, cell-cycle, Fluorescence Microscopy, Machine Learning
Tasks and Milestones
1. Adapt and Improve an existing automated segmentation and classification algorithm
2. Train a neural network
3. Statistical analysis
5. Report writing

